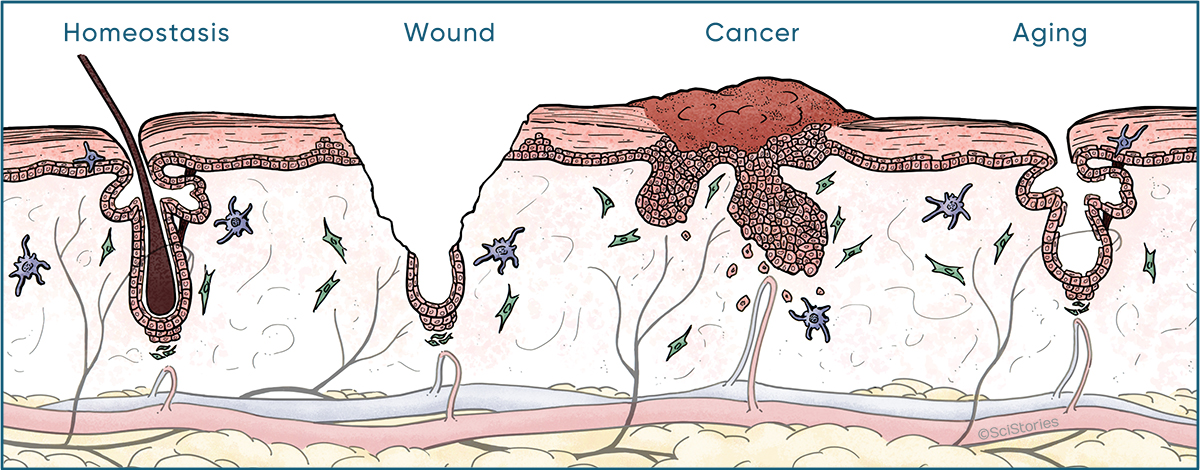
Defined by golden standards of long-term self-renewal and multi-lineage differentiation, stem cells (SCs) come in different flavors. In mammals, adult SCs are essential units to orchestrate postnatal remodeling and repair damage. In contrast to steady state, SCs in coping with stress often expand their fates and embark on behaviors distinct from their homeostatic patterns, known as plasticity. While plasticity is essential for organismal survival, its derailed regulation poses disease vulnerability to individuals, especially those undergoing prolonged stress. Under these scenarios, SCs are subjected to functional exhaustion frequently observed in aging, or malignant transformation that occurs in cancer. Research in the Ge lab applies principle of developmental biology in order to understand molecular mechanisms underlying SC plasticity, and how its deregulation leads to human diseases.
One of the key hypotheses we set out to test is this long postulated idea “cancer is a wound that never heals”. Parallels between wound repair and cancer have emerged in many contexts, begging questions in regards to their molecular origin and functional relevance. Mouse skin represents an excellent model to address these outstanding issues. Its SCs are well defined, abundant, and genetically tractable. Studying squamous cell carcinomas (SCCs) of the skin, resembling those in the head and neck, esophagus, lung and cervix which are highly deadly, we found SCs adopt a plastic phenotype so called “‘lineage infidelity”, entailing a wide spread co-expression of otherwise lineage restricted genes. Rather than a cancer specific phenomenon, lineage infidelity occurs transiently during normal wound repair and is functionally required for SCs to cope with stress. Lineage infidelity is hijacked by SCs of the SCCs where it is sustained to maintain malignancy. Mechanistically, it signifies a state where lineage genes are functionally re-wired from a homeostatic network into a stress-specific one, presumably in cooperation of stress-induced genes, at the level of chromatin and non-coding regulatory elements. Our most recent observations suggest lineage infidelity occurs across many diseases and stress conditions including aging and chronic wounds, therefore may represent a core mechanism SCs use to steer fate choices and a valuable therapeutic target in human patients.
For more details, please see Yejing’s recent talk at MDACC Enjoy Science. https://mediaplayer.mdanderson.org/video-full/9D2A4804-5FC4-40DC-97AA-82C229B3DBCE
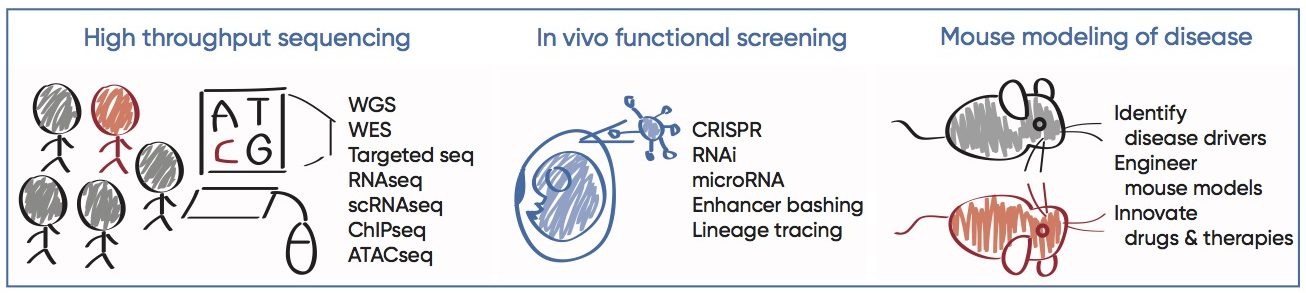
Entering the age of personalized medicine, large data sets are produced at unprecedented pace and depth. The second part of our research aims to exploit mouse genetics and functional genomics approaches, in the goal of bridging human disease association to gene function, and ultimately advancing disease treatments and patient care. We are particularly interested in interrogating the non-coding regulatory sequences as drivers of human diseases. So called the “genomic dark matter”, non-coding sequences comprise 98.5% of the human genome whose functional significance remains largely uncharted. Given our own observations of the dramatic remodeling of the non-coding regulatory elements underlying SC response in wound repair and cancer, it is intriguing to postulate their miss-regulation may lead to human diseases. Recent advance in high throughput sequencing and CRISPR technology provided unprecedented opportunities to gain mechanistic insights into these questions.
As proof of principle, we characterized one group of such non-coding products, microRNAs (miRNAs) — evolutionarily conserved, abundant, small non-coding RNAs that regulate gene expression at post-transcriptional level. Using ultrasound-guided in utero lentiviral microinjections, we carried out in vivo high throughput functional screen, through which we identified novel oncogenic miRNAs that drive the progression of skin SCCs. Recently, we’ve combined this system with CRISPR to achieve lineage specific and inducible knockout, overexpression, enhancer bashing, and lineage tracing to dissect molecular and cellular mechanisms underlying wound repair, cancer and aging. Such rapid, stable and tissue specific transgene integration enables functional genomics at a hitherto unparalleled speed and precision. Our long-term goal is to leverage the basic knowledge we learn from human data and mouse models to therapeutically benefit patients with degenerative and malignant diseases.